Work Package 6
Development of a framework for sharing and expanding capabilities in new breeding, phenotyping, and genomic technologies for farmed animals
A key focus of the EuroFAANG Research Infrastructure (RI) is to share and expand the use of new tools, concepts and technologies for farmed animal phenotyping and genomics.
A number of cutting-edge technologies are driving the field forward, and in Work Package 6 (WP6) we aim to develop a framework for stakeholders (e.g. breeding companies, researchers, farmers) across Europe to access these tools and, ultimately, to integrate and apply them to novel breeding and management practices. WP6 will therefore make an important contribution to achieving the overarching goal of EuroFAANG; to facilitate research and innovation aimed at producing healthier, more sustainable, farmed animals.
WP6 will also explore the potential to incorporate new and emerging farmed animal species (e.g. insects and marine invertebrates) into the EuroFAANG infrastructure. This will bring many new stakeholders into the field, who previously have not been represented in the EuroFAANG infrastructure.
Work Package 6 in a nutshell
WP6 aims to expand capabilities with new technologies in phenotyping and genomics, and to engage with stakeholders to promote the uptake of new tools and concepts thereby advancing the breeding and management of farmed animals across Europe.
Genomic technologies are tools that allow us to study the complete genetic code of farmed animals (their ‘genotypes’) in detail. These tools help to identify and exploit key genetic factors underpinning important traits that are relevant for breeding healthier animals in a more sustainable way. The main focus of WP6 will be on functional genomics-based precision breeding in collaboration with WP4 (in vitro cell systems) and WP5 (genome editing).
Check out a similar example on fish farming from the AQUAFAANG project.
Phenotyping technologies are the tools that allow us to measure key traits (phenotypes) that are important for improved breeding practices. Improving how we can link phenotypes in populations of animals to their genetic code (genotype), to improve prediction accuracy is one of the biggest challenges that animal breeders face. However, collecting this information at a sufficient scale is challenging. Linking genotype to phenotype in cellular systems such cells or organoids can provide an alternative to performing this work in whole organisms, minimising the use of live animals in research according to the 3R principles.
Using these cellular systems WP6 aims to develop strategies and resources to bridge the knowledge gap that currently exists between cell/organ- and organism-level information.
Another key objective of WP6 is to support the integration of genotype and phenotype information, and to link genetic markers with phenotypic traits. Such ‘genotype to phenotype’ (G2P) tools are essential to move towards more sustainable farming practices, and will require innovative new approaches, such as artificial intelligence (AI).
WP6 goals can be broadly divided into three main tasks:
Connecting an animal’s genotype to its phenotype requires detailed knowledge at all intermediate biological levels (e.g. genome – cell – organism – environment – population). This is a difficult task to achieve while preserving animal welfare and implementing the 3R principles to “refine, reduce and replace” the use of whole animals in research. Cellular in vitro models, including cell lines and organoids, provide the possibility to considerably reduce the use of whole animals for studies linking genotype to phenotype.
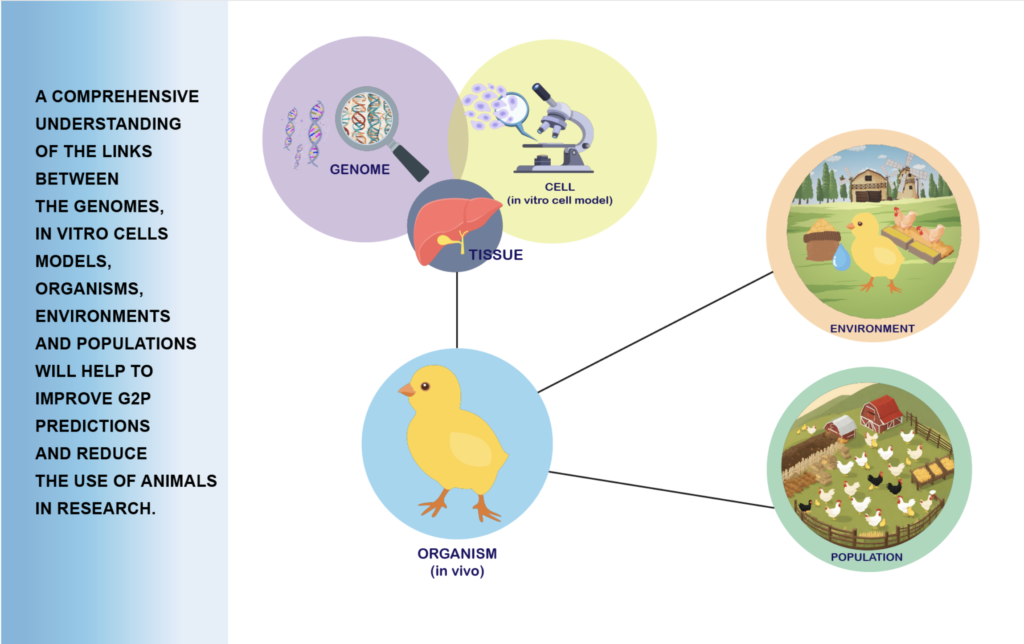
The aim of task 1 is to propose novel strategies based on cutting-edge technologies to bridge the phenotyping gap between in vitro cellular models and the ‘classical’ in vivo phenotyping approaches. This will provide a foundation for new approaches suited to study pressing topics in farmed animal production, particularly related to climate change (e.g. response to biotic and abiotic stress, e.g. heat) and sustainability (e.g. nutrient utilization).
In close collaboration with WP4, WP5 and WP7, we will establish an inventory of relevant infrastructures, networks, methods, and resources required to drive progress in this field (Tab 1). Experts will be engaged in all required fields including cell biology, genomics, and computational modelling to outline the current state of the art, identify needs, and propose a stepwise strategy towards bridging the gap between in vitro and in vivo approaches for G2P research.
WP6 will map potential stakeholders using genomic technologies for farmed animal breeding and management and work closely with these stakeholders to facilitate the uptake of G2P information in breeding programmes. To take full advantage of G2P information, stakeholders will need information on standard data formats and rules/procedures for data transfer (this aligns with WP3 efforts to establish a common data structure). We will create research activities focused on harnessing the power of genomic technologies to improve animal health and welfare, particularly in managing trade-offs between traits.
The increasing volume and resolution of data offer unprecedented opportunities for accurately linking genome information to phenotype (G2P research). However, limitations in current data analysis methods may restrict the extent to which this great potential is realised. For example, most methods look for simple linear relationships between phenotype and genotype, while in reality this relationship is driven by complex interactions between multiple genetic factors. Task 3 will explore the use of AI in G2P research to use genomic information for predicting traits. Such discoveries can contribute to more accurate predictions of the genetic merit of farmed animals, in particular for traits that have not been selected for (related to health, welfare, and behaviour). First, Task 3 will create a portfolio of laboratories and institutions in Europe that work towards the application of AI for G2P prediction. Second, we will create an overview of AI methods that have been tested for G2P prediction. Finally, we will identify training opportunities for industry partners to develop, test and transfer novel AI-based pipelines into their breeding programmes and management approaches.

Keep in touch!
EuroFAANG projects

GEroNIMO
Go to website
BovReg
Go to website
Rumigen
Go to website
GENE-SWitCH
Go to website
HoloRuminant
Go to website